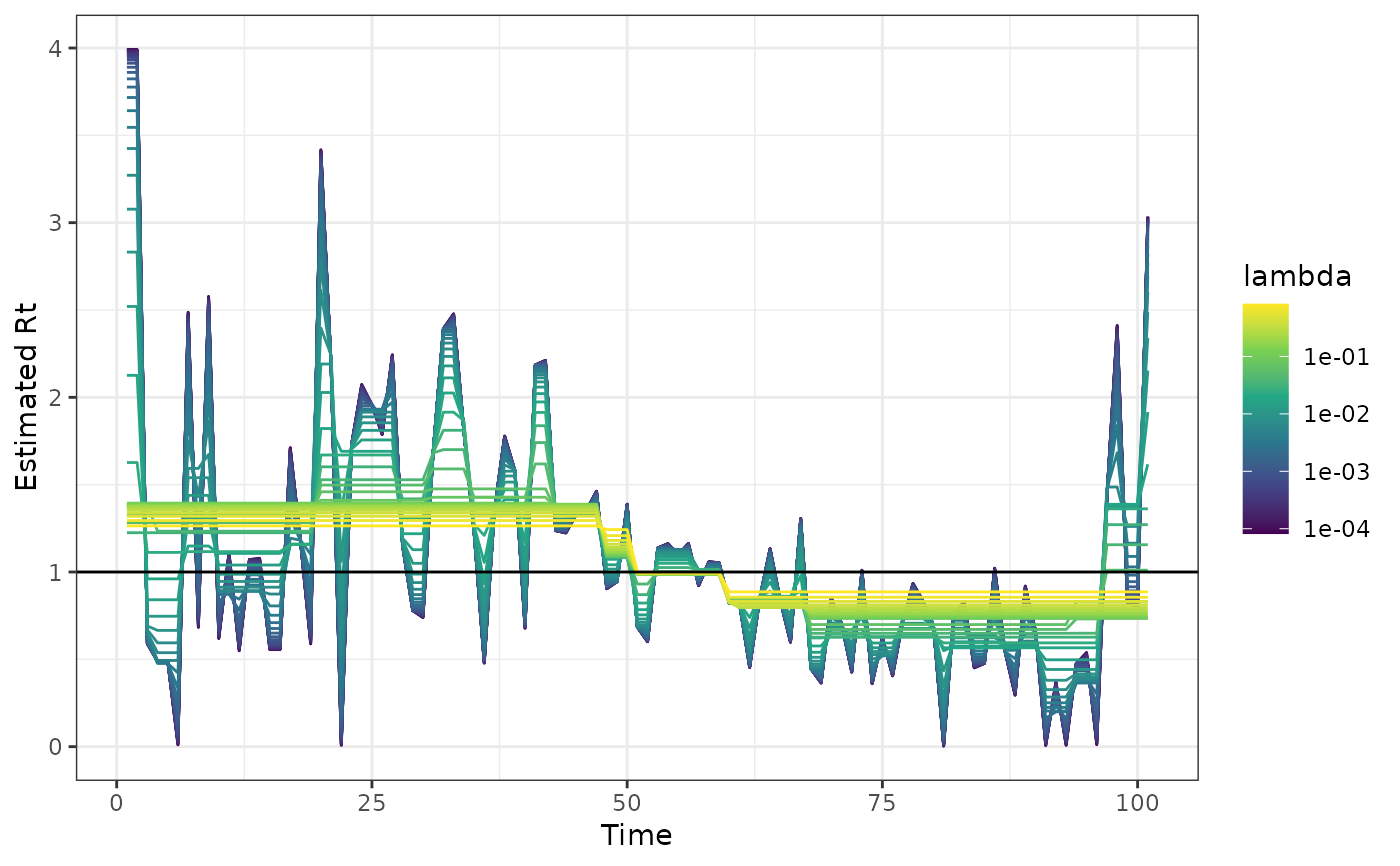
The Effective Reproduction Number \(R_t\) of an infectious disease can be estimated by solving the smoothness penalized Poisson regression (trend filtering) of the form:
$$\hat{\theta} = \arg\min_{\theta} \frac{1}{n} \sum_{i=1}^n (w_i e^{\theta_i} - y_i\theta_i) + \lambda\Vert D^{(k+1)}\theta\Vert_1, $$
where \(R_t = e^{\theta}\), \(y_i\) is the observed case count at day \(i\), \(w_i\) is the weighted past counts at day \(i\), \(\lambda\) is the smoothness penalty, and \(D^{(k+1)}\) is the \((k+1)\)-th order difference matrix.
Usage
estimate_rt(
observed_counts,
korder = 3L,
dist_gamma = c(2.5, 2.5),
x = 1:n,
lambda = NULL,
nsol = 50L,
delay_distn = NULL,
delay_distn_periodicity = NULL,
lambdamin = NULL,
lambdamax = NULL,
lambda_min_ratio = 1e-04,
maxiter = 1e+05,
init = configure_rt_admm()
)Arguments
- observed_counts
vector of the observed daily infection counts
- korder
Integer. Degree of the piecewise polynomial curve to be estimated. For example,
korder = 0corresponds to a piecewise constant curve.- dist_gamma
Vector of length 2. These are the shape and scale for the assumed serial interval distribution. Roughly, this distribution describes the probability of an infectious individual infecting someone else after some period of time after having become infectious. As in most literature, we assume that this interval follows a gamma distribution with some shape and scale.
- x
a vector of positions at which the counts have been observed. In an ideal case, we would observe data at regular intervals (e.g. daily or weekly) but this may not always be the case. May be numeric or Date.
- lambda
Vector. A user supplied sequence of tuning parameters which determines the balance between data fidelity and smoothness of the estimated Rt; larger
lambdaresults in a smoother estimate. The default,NULLresults in an automatic computation based onnlambda, the largest value oflambdathat would result in a maximally smooth estimate, andlambda_min_ratio. Supplying a value oflambdaoverrides this behaviour. It is likely better to supply a decreasing sequence oflambdavalues than a single (small) value. If supplied, the user-definedlambdasequence is automatically sorted in decreasing order.- nsol
Integer. The number of tuning parameters
lambdaat which to compute Rt.- delay_distn
in the case of a non-gamma delay distribution, a vector or matrix (or
Matrix::Matrix()) of delay probabilities may be passed here. For a vector, these will be coerced to sum to 1, and padded with 0 in the right tail if necessary. If a time-varying delay matrix, it must be lower-triangular. Each row will be silently coerced to sum to 1. See alsovignette("delay-distributions").- delay_distn_periodicity
Controls the relationship between the spacing of the computed delay distribution and the spacing of
x. In the default case,xwould be regular on the sequence1:length(observed_cases), and this would be 1. But ifxis aDateobject or spaced irregularly, the relationship becomes more complicated. For example, weekly data whenxis a date in the formYYYY-MM-DDrequires specifyingdelay_distn_periodicity = "1 week". Or ifobserved_caseswere reported on Monday, Wednesday, and Friday, thendelay_distn_periodicity = "1 day"would be most appropriate.- lambdamin
Optional value for the smallest
lambdato use. This should be greater than zero.- lambdamax
Optional value for the largest
lambdato use.- lambda_min_ratio
If neither
lambdanorlambdaminis specified, the program will generate a lambdamin by lambdamax * lambda_min_ratio. A multiplicative factor for the minimal lambda in thelambdasequence, wherelambdamin = lambda_min_ratio * lambdamax. A very small value will lead to the solutionRt = log(observed_counts). This argument has no effect if there is a user-definedlambdasequence.- maxiter
Integer. Maximum number of iterations for the estimation algorithm.
- init
a list of internal configuration parameters of class
rt_admm_configuration.
Value
An object with S3 class poisson_rt. Among the list components:
observed_countsthe observed daily infection counts.xa vector of positions at which the counts have been observed.weighted_past_countsthe weighted sum of past infection counts.Rtthe estimated effective reproduction rate. This is a matrix with each column corresponding to one value oflambda.lambdathe values oflambdaactually used in the algorithm.korderdegree of the estimated piecewise polynomial curve.dofdegrees of freedom of the estimated trend filtering problem.niterthe required number of iterations for each value oflambda.convergenceif number of iterations for each value oflambdais less than the maximum number of iterations for the estimation algorithm.
Examples
y <- c(1, rpois(100, dnorm(1:100, 50, 15) * 500 + 1))
out <- estimate_rt(y)
out
#>
#> Call: estimate_rt(observed_counts = y)
#>
#> Degree of the estimated piecewise polynomial curve: 3
#>
#> Summary of the 50 estimated models:
#>
#> lambda index approx_dof niterations
#> Max. 313.3978 1 4 13
#> 3rd Qu. 32.8476 13 4 84
#> Median 2.8529 26 6 525
#> 1st Qu. 0.2990 38 14 1581
#> Min. 0.0313 50 30 2330
#>
plot(out)
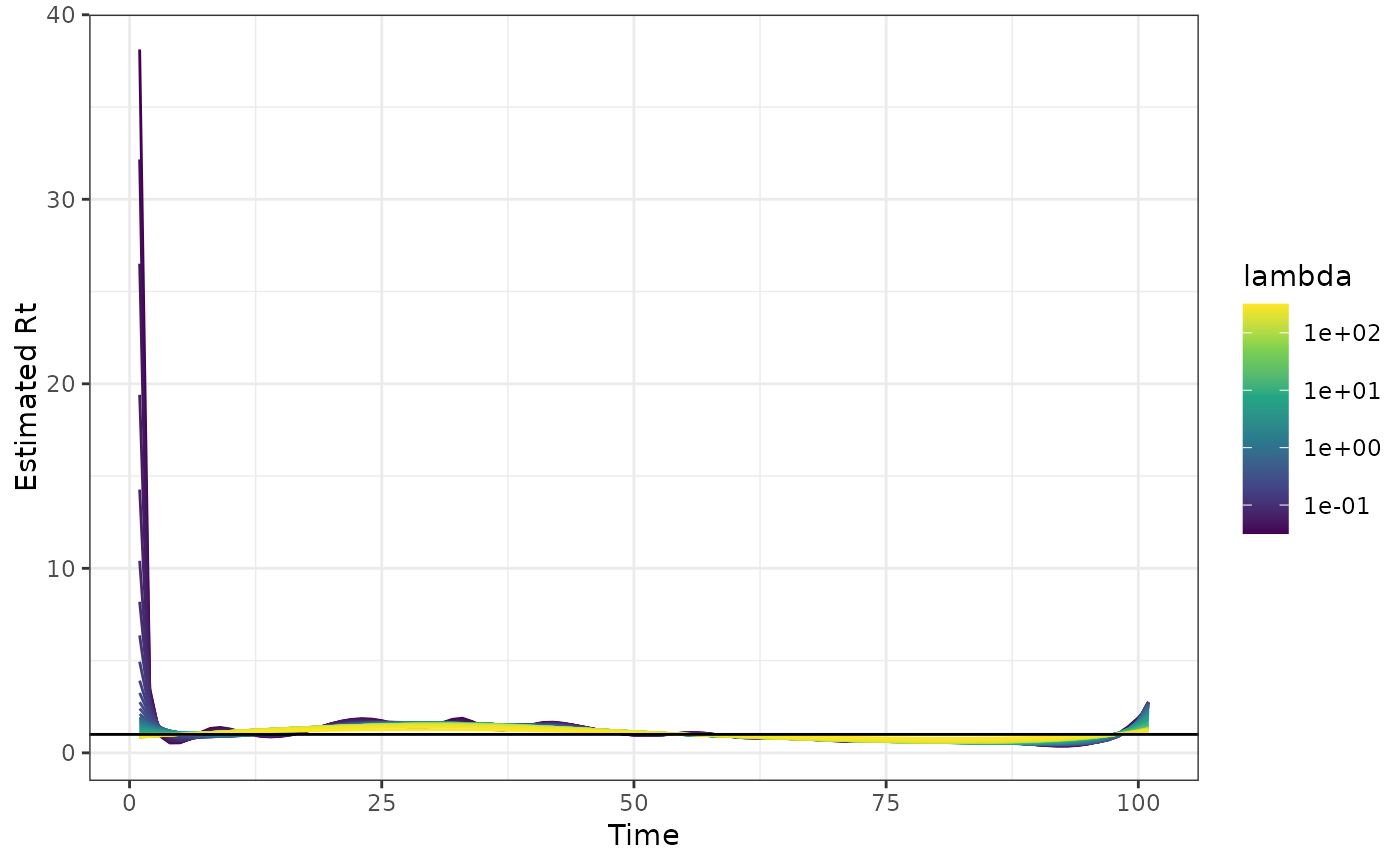 out0 <- estimate_rt(y, korder = 0L, nsol = 40)
out0
#>
#> Call: estimate_rt(observed_counts = y, korder = 0L, nsol = 40)
#>
#> Degree of the estimated piecewise polynomial curve: 0
#>
#> Summary of the 40 estimated models:
#>
#> lambda index approx_dof niterations
#> Max. 80.25910 1 1 0
#> 3rd Qu. 7.56577 11 9 0
#> Median 0.90318 20 54 0
#> 1st Qu. 0.08514 30 93 0
#> Min. 0.00803 40 96 0
#>
plot(out0)
out0 <- estimate_rt(y, korder = 0L, nsol = 40)
out0
#>
#> Call: estimate_rt(observed_counts = y, korder = 0L, nsol = 40)
#>
#> Degree of the estimated piecewise polynomial curve: 0
#>
#> Summary of the 40 estimated models:
#>
#> lambda index approx_dof niterations
#> Max. 80.25910 1 1 0
#> 3rd Qu. 7.56577 11 9 0
#> Median 0.90318 20 54 0
#> 1st Qu. 0.08514 30 93 0
#> Min. 0.00803 40 96 0
#>
plot(out0)