class: center, middle, inverse, title-slide # COVID-19 Modelling and Forecasting ## In the US and Canada<br/>A statistician’s pro(retro)spective ### Daniel J. McDonald ### 12 October 2021 --- class: middle .pull-left[.center[  ]] .pull-right[.center[   ]] --- ## Outline <hr> <br/> .right-column[ ### 1. Background on Epi Groups <br/> ### 2. Intro to standard epidemic models <br/> ### 3. Out-of-sample COVID forecasting <br/> ### 4. Gaps in forecasting <br/> ### 5. Some concluding thoughts ] --- layout: true ## BC COVID Modelling Group .left-column[ ### Personnel Prof. Dan Coombs Prof. Sally Otto Prof. Caroline Colijn Prof. Dean Karlen Prof. Eric Cytrynbaum Jens von Bergmann Others ] --- .right-column[ ### Goals * Volunteers interested and motivated by understanding COVID-19. * Began as a research group with invited speakers, paper discussion. * Focused on **understanding** local issues, providing a perspective on the situation that might help policy. * A focus on factual/counterfactual mathematical modeling. * Strong connection to local/national journalists (esp. Sally and Caroline). * Began releasing reports every other week in April. **Big emphasis on data issues/availability. Current conditions. News or PH statements.** ] --- .right-column[ ### Some recent topics * Is there evidence of "decoupling"? * Waning vaccine efficacy? [New evidence from BCCDC and INSQP](https://www.cbc.ca/news/health/canada-vaccine-effectiveness-data-delayed-doses-mixing-matching-covid-19-vaccines-1.6205993) * Why is there a surge in cases for `\(<\)` 10? Does testing explain it? * Why is there a testing gap in Alberta? * Why is it taking so long for sequencing results to hit the open-access portal? * What is the case for vaccinating 5-11 year olds? * What is the path from pandemic to endemic? And how soon? ] --- layout: false ## Delphi ### History * [Delphi](https://delphi.cmu.edu/) Formed in 2012, to "develop theory and practice of epi forecasting". * National US orientation. * Participated in annual CDC influenza forecasting challenges since 2013, and won several. * Awarded CDC National Center of Excellence for flu forecasting in 2019. * Pivoted in February 2020 to focus on supporting the US COVID-19 response, launched [COVIDcast project](https://delphi.cmu.edu/covidcast/). -- ### Goals **The full pipeline:** 1. Find, collect, disseminate the data we need. 1. Use the data to: model, forecast, nowcast, answer the big questions. 1. Create software for the community. 1. Advance research. 1. Influence policymakers. 1. Communicate broadly. --- layout: true ## Delphi personnel .left-column[  **Roni Rosenfeld** CMU MLD  **Ryan Tibshirani** CMU MLD + Stat ] --- --- .center[  ] --- .center[ <img src="more-delphi.png" width="30%" /><img src="even-more-delphi.png" width="30%" /> ] --- ### Recent efforts * Massive effort to collect and disseminate versioned data. * Website to display National-level indicators. * Survey on Facebook: ~24M responses, 12 waves of questions. * Creating Ensemble forecasts. * Work on many traditional stats research topics. (causal inference, nowcasting, CFR estimation, deconvolution, recalibration, backfill corrections, ...) * Software development. * Forecast production, evaluation, and scoring. --- layout: false ## To understand COVID-19, we need data .pull-left[ ### Canada <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M104 224H24c-13.255 0-24 10.745-24 24v240c0 13.255 10.745 24 24 24h80c13.255 0 24-10.745 24-24V248c0-13.255-10.745-24-24-24zM64 472c-13.255 0-24-10.745-24-24s10.745-24 24-24 24 10.745 24 24-10.745 24-24 24zM384 81.452c0 42.416-25.97 66.208-33.277 94.548h101.723c33.397 0 59.397 27.746 59.553 58.098.084 17.938-7.546 37.249-19.439 49.197l-.11.11c9.836 23.337 8.237 56.037-9.308 79.469 8.681 25.895-.069 57.704-16.382 74.757 4.298 17.598 2.244 32.575-6.148 44.632C440.202 511.587 389.616 512 346.839 512l-2.845-.001c-48.287-.017-87.806-17.598-119.56-31.725-15.957-7.099-36.821-15.887-52.651-16.178-6.54-.12-11.783-5.457-11.783-11.998v-213.77c0-3.2 1.282-6.271 3.558-8.521 39.614-39.144 56.648-80.587 89.117-113.111 14.804-14.832 20.188-37.236 25.393-58.902C282.515 39.293 291.817 0 312 0c24 0 72 8 72 81.452z"/></svg> Centralized Health Care <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M104 224H24c-13.255 0-24 10.745-24 24v240c0 13.255 10.745 24 24 24h80c13.255 0 24-10.745 24-24V248c0-13.255-10.745-24-24-24zM64 472c-13.255 0-24-10.745-24-24s10.745-24 24-24 24 10.745 24 24-10.745 24-24 24zM384 81.452c0 42.416-25.97 66.208-33.277 94.548h101.723c33.397 0 59.397 27.746 59.553 58.098.084 17.938-7.546 37.249-19.439 49.197l-.11.11c9.836 23.337 8.237 56.037-9.308 79.469 8.681 25.895-.069 57.704-16.382 74.757 4.298 17.598 2.244 32.575-6.148 44.632C440.202 511.587 389.616 512 346.839 512l-2.845-.001c-48.287-.017-87.806-17.598-119.56-31.725-15.957-7.099-36.821-15.887-52.651-16.178-6.54-.12-11.783-5.457-11.783-11.998v-213.77c0-3.2 1.282-6.271 3.558-8.521 39.614-39.144 56.648-80.587 89.117-113.111 14.804-14.832 20.188-37.236 25.393-58.902C282.515 39.293 291.817 0 312 0c24 0 72 8 72 81.452z"/></svg> Regularized reporting <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M104 224H24c-13.255 0-24 10.745-24 24v240c0 13.255 10.745 24 24 24h80c13.255 0 24-10.745 24-24V248c0-13.255-10.745-24-24-24zM64 472c-13.255 0-24-10.745-24-24s10.745-24 24-24 24 10.745 24 24-10.745 24-24 24zM384 81.452c0 42.416-25.97 66.208-33.277 94.548h101.723c33.397 0 59.397 27.746 59.553 58.098.084 17.938-7.546 37.249-19.439 49.197l-.11.11c9.836 23.337 8.237 56.037-9.308 79.469 8.681 25.895-.069 57.704-16.382 74.757 4.298 17.598 2.244 32.575-6.148 44.632C440.202 511.587 389.616 512 346.839 512l-2.845-.001c-48.287-.017-87.806-17.598-119.56-31.725-15.957-7.099-36.821-15.887-52.651-16.178-6.54-.12-11.783-5.457-11.783-11.998v-213.77c0-3.2 1.282-6.271 3.558-8.521 39.614-39.144 56.648-80.587 89.117-113.111 14.804-14.832 20.188-37.236 25.393-58.902C282.515 39.293 291.817 0 312 0c24 0 72 8 72 81.452z"/></svg> <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:orange;overflow:visible;position:relative;"><path d="M440.5 88.5l-52 52L415 167c9.4 9.4 9.4 24.6 0 33.9l-17.4 17.4c11.8 26.1 18.4 55.1 18.4 85.6 0 114.9-93.1 208-208 208S0 418.9 0 304 93.1 96 208 96c30.5 0 59.5 6.6 85.6 18.4L311 97c9.4-9.4 24.6-9.4 33.9 0l26.5 26.5 52-52 17.1 17zM500 60h-24c-6.6 0-12 5.4-12 12s5.4 12 12 12h24c6.6 0 12-5.4 12-12s-5.4-12-12-12zM440 0c-6.6 0-12 5.4-12 12v24c0 6.6 5.4 12 12 12s12-5.4 12-12V12c0-6.6-5.4-12-12-12zm33.9 55l17-17c4.7-4.7 4.7-12.3 0-17-4.7-4.7-12.3-4.7-17 0l-17 17c-4.7 4.7-4.7 12.3 0 17 4.8 4.7 12.4 4.7 17 0zm-67.8 0c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17zm67.8 34c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17zM112 272c0-35.3 28.7-64 64-64 8.8 0 16-7.2 16-16s-7.2-16-16-16c-52.9 0-96 43.1-96 96 0 8.8 7.2 16 16 16s16-7.2 16-16z"/></svg> Huge emphasis on privacy <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:orange;overflow:visible;position:relative;"><path d="M440.5 88.5l-52 52L415 167c9.4 9.4 9.4 24.6 0 33.9l-17.4 17.4c11.8 26.1 18.4 55.1 18.4 85.6 0 114.9-93.1 208-208 208S0 418.9 0 304 93.1 96 208 96c30.5 0 59.5 6.6 85.6 18.4L311 97c9.4-9.4 24.6-9.4 33.9 0l26.5 26.5 52-52 17.1 17zM500 60h-24c-6.6 0-12 5.4-12 12s5.4 12 12 12h24c6.6 0 12-5.4 12-12s-5.4-12-12-12zM440 0c-6.6 0-12 5.4-12 12v24c0 6.6 5.4 12 12 12s12-5.4 12-12V12c0-6.6-5.4-12-12-12zm33.9 55l17-17c4.7-4.7 4.7-12.3 0-17-4.7-4.7-12.3-4.7-17 0l-17 17c-4.7 4.7-4.7 12.3 0 17 4.8 4.7 12.4 4.7 17 0zm-67.8 0c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17zm67.8 34c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17zM112 272c0-35.3 28.7-64 64-64 8.8 0 16-7.2 16-16s-7.2-16-16-16c-52.9 0-96 43.1-96 96 0 8.8 7.2 16 16 16s16-7.2 16-16z"/></svg> Relatively underfunded. <svg aria-hidden="true" role="img" viewBox="0 0 640 512" style="height:2em;width:2.5em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M621.16 54.46C582.37 38.19 543.55 32 504.75 32c-123.17-.01-246.33 62.34-369.5 62.34-30.89 0-61.76-3.92-92.65-13.72-3.47-1.1-6.95-1.62-10.35-1.62C15.04 79 0 92.32 0 110.81v317.26c0 12.63 7.23 24.6 18.84 29.46C57.63 473.81 96.45 480 135.25 480c123.17 0 246.34-62.35 369.51-62.35 30.89 0 61.76 3.92 92.65 13.72 3.47 1.1 6.95 1.62 10.35 1.62 17.21 0 32.25-13.32 32.25-31.81V83.93c-.01-12.64-7.24-24.6-18.85-29.47zM48 132.22c20.12 5.04 41.12 7.57 62.72 8.93C104.84 170.54 79 192.69 48 192.69v-60.47zm0 285v-47.78c34.37 0 62.18 27.27 63.71 61.4-22.53-1.81-43.59-6.31-63.71-13.62zM320 352c-44.19 0-80-42.99-80-96 0-53.02 35.82-96 80-96s80 42.98 80 96c0 53.03-35.83 96-80 96zm272 27.78c-17.52-4.39-35.71-6.85-54.32-8.44 5.87-26.08 27.5-45.88 54.32-49.28v57.72zm0-236.11c-30.89-3.91-54.86-29.7-55.81-61.55 19.54 2.17 38.09 6.23 55.81 12.66v48.89z"/></svg> goes to the "right places" ] .pull-right[ ### USA <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:orange;overflow:visible;position:relative;"><path d="M440.5 88.5l-52 52L415 167c9.4 9.4 9.4 24.6 0 33.9l-17.4 17.4c11.8 26.1 18.4 55.1 18.4 85.6 0 114.9-93.1 208-208 208S0 418.9 0 304 93.1 96 208 96c30.5 0 59.5 6.6 85.6 18.4L311 97c9.4-9.4 24.6-9.4 33.9 0l26.5 26.5 52-52 17.1 17zM500 60h-24c-6.6 0-12 5.4-12 12s5.4 12 12 12h24c6.6 0 12-5.4 12-12s-5.4-12-12-12zM440 0c-6.6 0-12 5.4-12 12v24c0 6.6 5.4 12 12 12s12-5.4 12-12V12c0-6.6-5.4-12-12-12zm33.9 55l17-17c4.7-4.7 4.7-12.3 0-17-4.7-4.7-12.3-4.7-17 0l-17 17c-4.7 4.7-4.7 12.3 0 17 4.8 4.7 12.4 4.7 17 0zm-67.8 0c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17zm67.8 34c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17zM112 272c0-35.3 28.7-64 64-64 8.8 0 16-7.2 16-16s-7.2-16-16-16c-52.9 0-96 43.1-96 96 0 8.8 7.2 16 16 16s16-7.2 16-16z"/></svg> Decentralized Health Care <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:orange;overflow:visible;position:relative;"><path d="M440.5 88.5l-52 52L415 167c9.4 9.4 9.4 24.6 0 33.9l-17.4 17.4c11.8 26.1 18.4 55.1 18.4 85.6 0 114.9-93.1 208-208 208S0 418.9 0 304 93.1 96 208 96c30.5 0 59.5 6.6 85.6 18.4L311 97c9.4-9.4 24.6-9.4 33.9 0l26.5 26.5 52-52 17.1 17zM500 60h-24c-6.6 0-12 5.4-12 12s5.4 12 12 12h24c6.6 0 12-5.4 12-12s-5.4-12-12-12zM440 0c-6.6 0-12 5.4-12 12v24c0 6.6 5.4 12 12 12s12-5.4 12-12V12c0-6.6-5.4-12-12-12zm33.9 55l17-17c4.7-4.7 4.7-12.3 0-17-4.7-4.7-12.3-4.7-17 0l-17 17c-4.7 4.7-4.7 12.3 0 17 4.8 4.7 12.4 4.7 17 0zm-67.8 0c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17zm67.8 34c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17zM112 272c0-35.3 28.7-64 64-64 8.8 0 16-7.2 16-16s-7.2-16-16-16c-52.9 0-96 43.1-96 96 0 8.8 7.2 16 16 16s16-7.2 16-16z"/></svg> Massively irregular reporting <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M104 224H24c-13.255 0-24 10.745-24 24v240c0 13.255 10.745 24 24 24h80c13.255 0 24-10.745 24-24V248c0-13.255-10.745-24-24-24zM64 472c-13.255 0-24-10.745-24-24s10.745-24 24-24 24 10.745 24 24-10.745 24-24 24zM384 81.452c0 42.416-25.97 66.208-33.277 94.548h101.723c33.397 0 59.397 27.746 59.553 58.098.084 17.938-7.546 37.249-19.439 49.197l-.11.11c9.836 23.337 8.237 56.037-9.308 79.469 8.681 25.895-.069 57.704-16.382 74.757 4.298 17.598 2.244 32.575-6.148 44.632C440.202 511.587 389.616 512 346.839 512l-2.845-.001c-48.287-.017-87.806-17.598-119.56-31.725-15.957-7.099-36.821-15.887-52.651-16.178-6.54-.12-11.783-5.457-11.783-11.998v-213.77c0-3.2 1.282-6.271 3.558-8.521 39.614-39.144 56.648-80.587 89.117-113.111 14.804-14.832 20.188-37.236 25.393-58.902C282.515 39.293 291.817 0 312 0c24 0 72 8 72 81.452z"/></svg> <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:orange;overflow:visible;position:relative;"><path d="M440.5 88.5l-52 52L415 167c9.4 9.4 9.4 24.6 0 33.9l-17.4 17.4c11.8 26.1 18.4 55.1 18.4 85.6 0 114.9-93.1 208-208 208S0 418.9 0 304 93.1 96 208 96c30.5 0 59.5 6.6 85.6 18.4L311 97c9.4-9.4 24.6-9.4 33.9 0l26.5 26.5 52-52 17.1 17zM500 60h-24c-6.6 0-12 5.4-12 12s5.4 12 12 12h24c6.6 0 12-5.4 12-12s-5.4-12-12-12zM440 0c-6.6 0-12 5.4-12 12v24c0 6.6 5.4 12 12 12s12-5.4 12-12V12c0-6.6-5.4-12-12-12zm33.9 55l17-17c4.7-4.7 4.7-12.3 0-17-4.7-4.7-12.3-4.7-17 0l-17 17c-4.7 4.7-4.7 12.3 0 17 4.8 4.7 12.4 4.7 17 0zm-67.8 0c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17zm67.8 34c-4.7-4.7-12.3-4.7-17 0-4.7 4.7-4.7 12.3 0 17l17 17c4.7 4.7 12.3 4.7 17 0 4.7-4.7 4.7-12.3 0-17l-17-17zM112 272c0-35.3 28.7-64 64-64 8.8 0 16-7.2 16-16s-7.2-16-16-16c-52.9 0-96 43.1-96 96 0 8.8 7.2 16 16 16s16-7.2 16-16z"/></svg> Occasional emphasis on "privacy" <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:1em;width:1em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M104 224H24c-13.255 0-24 10.745-24 24v240c0 13.255 10.745 24 24 24h80c13.255 0 24-10.745 24-24V248c0-13.255-10.745-24-24-24zM64 472c-13.255 0-24-10.745-24-24s10.745-24 24-24 24 10.745 24 24-10.745 24-24 24zM384 81.452c0 42.416-25.97 66.208-33.277 94.548h101.723c33.397 0 59.397 27.746 59.553 58.098.084 17.938-7.546 37.249-19.439 49.197l-.11.11c9.836 23.337 8.237 56.037-9.308 79.469 8.681 25.895-.069 57.704-16.382 74.757 4.298 17.598 2.244 32.575-6.148 44.632C440.202 511.587 389.616 512 346.839 512l-2.845-.001c-48.287-.017-87.806-17.598-119.56-31.725-15.957-7.099-36.821-15.887-52.651-16.178-6.54-.12-11.783-5.457-11.783-11.998v-213.77c0-3.2 1.282-6.271 3.558-8.521 39.614-39.144 56.648-80.587 89.117-113.111 14.804-14.832 20.188-37.236 25.393-58.902C282.515 39.293 291.817 0 312 0c24 0 72 8 72 81.452z"/></svg> Massively overfunded. <svg aria-hidden="true" role="img" viewBox="0 0 640 512" style="height:2em;width:2.5em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M621.16 54.46C582.37 38.19 543.55 32 504.75 32c-123.17-.01-246.33 62.34-369.5 62.34-30.89 0-61.76-3.92-92.65-13.72-3.47-1.1-6.95-1.62-10.35-1.62C15.04 79 0 92.32 0 110.81v317.26c0 12.63 7.23 24.6 18.84 29.46C57.63 473.81 96.45 480 135.25 480c123.17 0 246.34-62.35 369.51-62.35 30.89 0 61.76 3.92 92.65 13.72 3.47 1.1 6.95 1.62 10.35 1.62 17.21 0 32.25-13.32 32.25-31.81V83.93c-.01-12.64-7.24-24.6-18.85-29.47zM48 132.22c20.12 5.04 41.12 7.57 62.72 8.93C104.84 170.54 79 192.69 48 192.69v-60.47zm0 285v-47.78c34.37 0 62.18 27.27 63.71 61.4-22.53-1.81-43.59-6.31-63.71-13.62zM320 352c-44.19 0-80-42.99-80-96 0-53.02 35.82-96 80-96s80 42.98 80 96c0 53.03-35.83 96-80 96zm272 27.78c-17.52-4.39-35.71-6.85-54.32-8.44 5.87-26.08 27.5-45.88 54.32-49.28v57.72zm0-236.11c-30.89-3.91-54.86-29.7-55.81-61.55 19.54 2.17 38.09 6.23 55.81 12.66v48.89z"/></svg> <svg aria-hidden="true" role="img" viewBox="0 0 640 512" style="height:2em;width:2.5em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M621.16 54.46C582.37 38.19 543.55 32 504.75 32c-123.17-.01-246.33 62.34-369.5 62.34-30.89 0-61.76-3.92-92.65-13.72-3.47-1.1-6.95-1.62-10.35-1.62C15.04 79 0 92.32 0 110.81v317.26c0 12.63 7.23 24.6 18.84 29.46C57.63 473.81 96.45 480 135.25 480c123.17 0 246.34-62.35 369.51-62.35 30.89 0 61.76 3.92 92.65 13.72 3.47 1.1 6.95 1.62 10.35 1.62 17.21 0 32.25-13.32 32.25-31.81V83.93c-.01-12.64-7.24-24.6-18.85-29.47zM48 132.22c20.12 5.04 41.12 7.57 62.72 8.93C104.84 170.54 79 192.69 48 192.69v-60.47zm0 285v-47.78c34.37 0 62.18 27.27 63.71 61.4-22.53-1.81-43.59-6.31-63.71-13.62zM320 352c-44.19 0-80-42.99-80-96 0-53.02 35.82-96 80-96s80 42.98 80 96c0 53.03-35.83 96-80 96zm272 27.78c-17.52-4.39-35.71-6.85-54.32-8.44 5.87-26.08 27.5-45.88 54.32-49.28v57.72zm0-236.11c-30.89-3.91-54.86-29.7-55.81-61.55 19.54 2.17 38.09 6.23 55.81 12.66v48.89z"/></svg> <svg aria-hidden="true" role="img" viewBox="0 0 640 512" style="height:2em;width:2.5em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:green;overflow:visible;position:relative;"><path d="M621.16 54.46C582.37 38.19 543.55 32 504.75 32c-123.17-.01-246.33 62.34-369.5 62.34-30.89 0-61.76-3.92-92.65-13.72-3.47-1.1-6.95-1.62-10.35-1.62C15.04 79 0 92.32 0 110.81v317.26c0 12.63 7.23 24.6 18.84 29.46C57.63 473.81 96.45 480 135.25 480c123.17 0 246.34-62.35 369.51-62.35 30.89 0 61.76 3.92 92.65 13.72 3.47 1.1 6.95 1.62 10.35 1.62 17.21 0 32.25-13.32 32.25-31.81V83.93c-.01-12.64-7.24-24.6-18.85-29.47zM48 132.22c20.12 5.04 41.12 7.57 62.72 8.93C104.84 170.54 79 192.69 48 192.69v-60.47zm0 285v-47.78c34.37 0 62.18 27.27 63.71 61.4-22.53-1.81-43.59-6.31-63.71-13.62zM320 352c-44.19 0-80-42.99-80-96 0-53.02 35.82-96 80-96s80 42.98 80 96c0 53.03-35.83 96-80 96zm272 27.78c-17.52-4.39-35.71-6.85-54.32-8.44 5.87-26.08 27.5-45.88 54.32-49.28v57.72zm0-236.11c-30.89-3.91-54.86-29.7-55.81-61.55 19.54 2.17 38.09 6.23 55.81 12.66v48.89z"/></svg> goes to the all the wrong places ] <hr> -- <br/> .center[ .large[**Data Collection and Quality Is A Major Issue**] [Wash Post 9/29: "Messy, incomplete U.S. data hobbles pandemic response"](https://www.washingtonpost.com/health/2021/09/30/inadequate-us-data-pandemic-response/?utm_campaign=wp_main&utm_medium=social&utm_source=facebook&fbclid=IwAR1JgXZOqqUVmbhI2htn3JLsEiixunXlZEr5wHr5euFCNcJdYBVCL9cn9lA) ] -- .center[.large[**The Necessary Data Depends on your Goals**]] --- class: inverse, middle, center # Standard epidemic models --- layout: true ## SIR-type (compartmental) models - Stochastic Version --- (Borrowed heavily from [Cosma Shalizi](http://www.stat.cmu.edu/~cshalizi/dst/20/lectures/20/lecture-20.html), Ryan Tibshirani, [Dean Karlen](https://pypm.github.io/home/docs/studies/reports/Characterizing_spread.pdf)) .pull-left[ Suppose each of N people in a bucket at time t: 1. .large[.primary[Susceptible(t)]] : not sick, but could get sick 1. .large[.secondary[Infected(t)]] : sick, can make others sick 1. .large[.tertiary[Removed(t)]] : either recovered or dead; not sick, can't get sick, can't make anyone sick ] .pull-right[ <div id="htmlwidget-4e06e4f80c8de896e434" style="width:100%;height:144px;" class="widgetframe html-widget"></div> <script type="application/json" data-for="htmlwidget-4e06e4f80c8de896e434">{"x":{"url":"index_files/figure-html//widgets/widget_sir-dag.html","options":{"xdomain":"*","allowfullscreen":false,"lazyload":false}},"evals":[],"jsHooks":[]}</script> ] <hr> **Assume**: During period h, each .primary[S] meets kh people. **Assume**: Prob( .primary[S] meets .secondary[I] and becomes .secondary[I] ) = c. .fourth-color[So]: Prob( .primary[S] `\(\rightarrow\)` .secondary[I] ) = `\(1 - (1 - c I(t) / N )^{hk} \approx kchI(t) / N\)` .fourth-color[So]: New .secondary[I] in time h `\(\sim Binom(S(t),\ kchI(t) / N)\)` **Assume**: Prob( .secondary[I] `\(\rightarrow\)` .tertiary[R]) = `\(\gamma h\)` .fourth-color[So]: new removals in time h `\(\sim Binom(I(t),\ \gamma h)\)` --- .pull-left-wide[ ### Over-all equations: `\begin{aligned} C(t+h) & = \mathrm{Binom}\left(S(t),\ \frac{\beta}{N} h I(t)\right)\\ D(t+h) & = \mathrm{Binom}\left(I(t),\ \gamma h\right)\\ S(t+h) & = S(t) - C(t+h)\\ I(t+h) & = I(t) + C(t+h) - D(t+h)\\ R(t+h) & = R(t) + D(t+h) \end{aligned}` ### In the deterministic limit, `\(N\rightarrow\infty,\ h\rightarrow 0\)` `\begin{aligned} N &= S(0) + I(0) + R(0)\\ \frac{dS}{dt} & = -\frac{\beta}{N} S(t)I(t)\\ \frac{dI}{dt} & = \frac{\beta}{N} I(t)S(t) - \gamma I(t)\\ \frac{dR}{dt} & = \gamma I(t) \end{aligned}` **"_the_ SIR model"** is often ambiguous between these ] .pull-right-narrow[ <div id="htmlwidget-d388c6527ad2212105bf" style="width:100%;height:252px;" class="widgetframe html-widget"></div> <script type="application/json" data-for="htmlwidget-d388c6527ad2212105bf">{"x":{"url":"index_files/figure-html//widgets/widget_sir-dag-again.html","options":{"xdomain":"*","allowfullscreen":false,"lazyload":false}},"evals":[],"jsHooks":[]}</script> ] --- layout: false ## Data issues - **Ideally** we'd see .large[.primary[S], .secondary[I], .tertiary[R]] at all times - Easier to observe new infections, .large[.secondary[I(t+h) - I(t)]] - Removals by death are easier to observe than removals by recovery, so we mostly see .large[.tertiary[(R(t+h) - R(t))] × (death rate)] - The interval between measurements, say `\(\Delta\)`, is often `\(\gg h\)` - Measuring .large[.secondary[I(t)]] and .large[.tertiary[R(t)]] (or their rates of change) is hard + testing/reporting is sporadic and error prone + Need to model test error (false positives, false negatives) _and_ who gets tested + Need to model lag between testing and reporting - Parameters (especially, `\(\beta\)`) change during the epidemic + Changing behavior, changing policy, environmental factors, vaccines, variants, ... --- layout: true ## Connecting to Data --- - Likelihood calculations are straightforward if we can measure .large[.secondary[I(t)], .tertiary[R(t)]] at all times 0, h, 2h, … T - Or .large[.secondary[I(0)]], .large[.tertiary[R(0)]] and all the increments .large[.secondary[I(t+h) - I(t)], .tertiary[R(t+h) - R(t)]] - Still have to optimize numerically - Likelihood calculations already become difficult if the time between observations `\(\Delta \gg h\)` + Generally, `\(\Delta \approx\)` 1 day + In principle, this just defines another Markov process, with a longer interval `\(\Delta\)` between steps, but to get the likelihood of a `\(\Delta\)` step we have to sum over all possible paths of `\(h\)` steps adding up to it - Other complications if we don't observe all the compartments, and/or have a lot of noise in our observations + We don't and we do. --- .pull-left[ - Often more tractable to avoid likelihood (Conditional least squares, simulation-based inference) - Intrinsic issue: Initially, everything just looks exponential + So it's hard to discriminate between distinct models + So even assuming an SIR model, it's easier to estimate `\(\beta - \gamma\)` than `\((\beta, \gamma)\)` or `\(\beta/\gamma\)` - Can sometimes **calibrate** or fix the parameters based on other sources + E.g., `\(1/\gamma =\)` average time someone is infectious, which could be determined from clinical studies / observations ] .pull-right[ <blockquote class="twitter-tweet"><p lang="en" dir="ltr">I have been thinking about how different people interpret data differently. And made this xkcd style graphic to illustrate this. <a href="https://t.co/a8LvlmZxT7">pic.twitter.com/a8LvlmZxT7</a></p>— Jens von Bergmann (@vb_jens) <a href="https://twitter.com/vb_jens/status/1372251931444350976?ref_src=twsrc%5Etfw">March 17, 2021</a></blockquote> ] --- layout: true ## BC COVID Compartmental Models .pull-left[.midi[ Two main models - Caroline Colijn (and team) [COVIDSEIR](https://seananderson.github.io/covidseir/index.html) * Uses the ODE version * Extra compartments for social distancing, others * Severity "ramp" on `\(\beta\)` * Likehood evaluation using R / Stan * Versions for age, a number of other things - Dean Karlen [pyPM](https://pypm.github.io/home/) * Discrete difference version * Extra compartments for VOCs, Hospitalizations, deaths, ICU, others * Estimated with Conditional Least Squares * Includes "Case Injections", estimated and forced changes in `\(\beta\)` * (Really a whole `python` package for creating compartmental models) - Both model reporting delays, seasonality - Other Bells and Whistles I'm not doing justice ]] --- .pull-right[  .small[.center[Source: [Dean Karlen](https://pypm.github.io/home/docs/studies/bc20211004/) 4 October model fit to all BC]] ] --- .pull-right[ 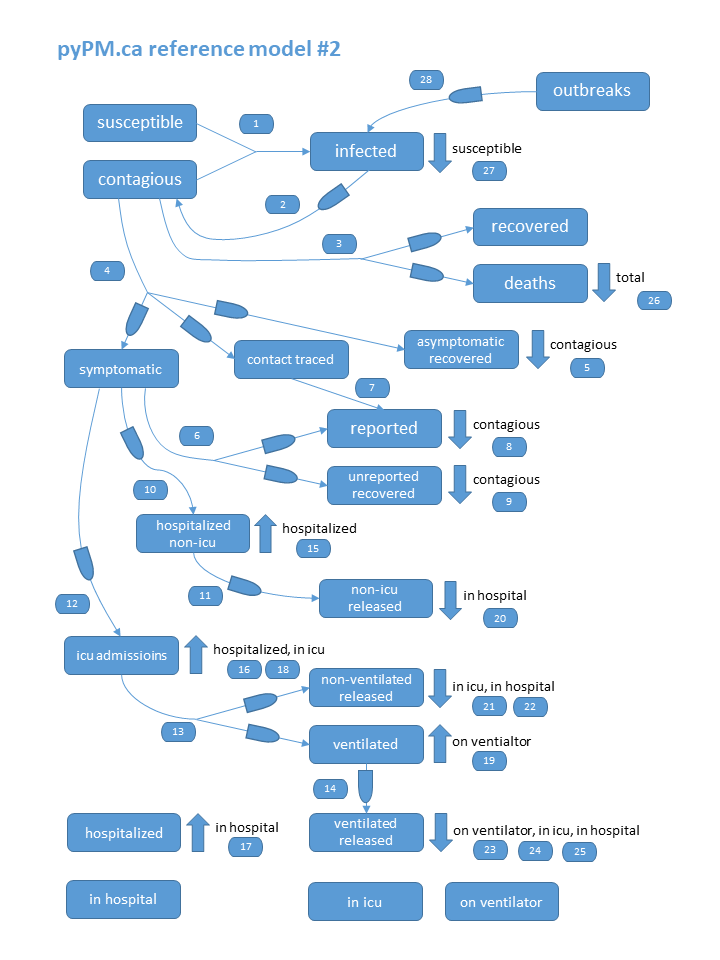 ] --- layout: false ## These models fit well "in sample" .pull-left[ * Track observed cases closely (they should) * Can provide nuanced policy advice on some topics * Many questions depend on modulating `\(\beta\)` 1. What happens if we lock down? 2. What happens if we mask? 3. What happens if we have school online? 4. Vaccine passport? * Vaccination modeling is easier, directly removes susceptibles * What about out-of-sample? ] -- .pull-right[.center[ <svg aria-hidden="true" role="img" viewBox="0 0 512 512" style="height:20em;width:20em;vertical-align:-0.125em;margin-left:auto;margin-right:auto;font-size:inherit;fill:orange;overflow:visible;position:relative;"><path d="M504 256c0 136.997-111.043 248-248 248S8 392.997 8 256C8 119.083 119.043 8 256 8s248 111.083 248 248zm-248 50c-25.405 0-46 20.595-46 46s20.595 46 46 46 46-20.595 46-46-20.595-46-46-46zm-43.673-165.346l7.418 136c.347 6.364 5.609 11.346 11.982 11.346h48.546c6.373 0 11.635-4.982 11.982-11.346l7.418-136c.375-6.874-5.098-12.654-11.982-12.654h-63.383c-6.884 0-12.356 5.78-11.981 12.654z"/></svg> ]] --- background-image: url("covidseir-fcast.pdf") background-position: center background-size: contain --- background-image: url("pypm-fcast.pdf") background-position: center background-size: contain --- class: inverse, middle, center # Out-of-sample COVID Forecasting --- ## The task .pull-left-wide[ * Every week, submit forecasts to [COVID-19 **Forecast**Hub](https://covid19forecasthub.org) * The first forecasts were submitted on 20 July 20 2020 * Originally "State Deaths" * Added "County Cases" a bit later, and eventually "Hospitalizations" * Hub also collects (now) a few other targets ] .pull-right-narrow[  ] -- ### The goal * Forecasts from many teams are combined into an "ensemble" * The ensemble generally has the best performance, and is the official [CDC Forecast](https://covid.cdc.gov/covid-data-tracker/#forecasting) * Some members of ForecastHub and Delphi collaborate on [Ensemble creation](https://forecasters.org/blog/2020/10/28/comparing-ensemble-approaches-for-short-term-probabilistic-covid-19-forecasts-in-the-u-s/) --- ## The Forecasters .pull-left[ * SIR, Agent-based models, ML models * Dean submits his * Delphi submitted simple ML until July ] .pull-right[ * "point" forecasts and "quantiles" * Predict 1-4 epiweeks ahead * Some teams produce longer-term forecasts ... ] <img src="index_files/figure-html/unnamed-chunk-2-1.svg" width="100%" /> --- ## Forecast evaluation [Weighted Interval Score (Bracher et al., 2000)](https://arxiv.org/abs/2005.12881) .pull-left[ * Calculated for each target (forecast date, location, horizon) * For each Interval: 1. Width of interval. 1. Under prediction (AE to the top) 1. Over prediction (AE to the bottom) * Weighted average using probability content * Mathematically equivalent to an average of quantile losses * Discrete approximation of CRPS ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-3-1.svg" width="100%" /> ] --- ## Comparing across space and time .pull-left[ * Error proportional to cases * Large right skew * "Adjust" by scaling to a Baseline * Baseline is flat line + residual quantiles <img src="index_files/figure-html/wis-map-1.svg" width="100%" /> ] .pull-right[ <img src="index_files/figure-html/wis-densities-1.svg" width="100%" /> ] --- ## Performance over time (Incident Deaths) <img src="index_files/figure-html/death-scores-all-1.svg" width="100%" /> --- ## Overall performance (Incident Deaths) #### All forecasters with at least 4 months of submissions <img src="index_files/figure-html/overall-death-1.svg" width="100%" /> --- ## What about Incident Cases? <img src="index_files/figure-html/overall-cases-1.svg" width="100%" /> --- class: middle, center, inverse # Gaps in forecasting --- ## Predicting surges in Florida <iframe src="https://ubc-stat-dajmcdon.shinyapps.io/fl-fcast-visualizer/" width="100%" height="700px" style="border:none"></iframe> --- ## PI Coverage <img src="index_files/figure-html/coverage-plot-1.svg" width="100%" /> --- layout: true ## The terrible case of Ohio --- <img src="index_files/figure-html/plot-oh-1.svg" width="100%" /> --- <img src="index_files/figure-html/bad-forecast-1.svg" width="100%" /> --- layout: false class: inverse, middle, center # Forecasting lessons --- layout: true ## Forecasting lessons --- Simple models [M, Bien, Green, Hu, ..., Tibshirani](http://medrxiv.org/content/10.1101/2021.06.22.21259346v1) <img src="index_files/figure-html/add-hrr-1.svg" width="100%" /> --- Human in the loop <img src="manual-corrections.png" width="100%" height="90%" /> --- Human in the loop <img src="quality-control.png" width="90%" /> --- layout:false class: middle, inverse, center # Thoughts and thanks --- ## Thoughts without pictures .pull-left[ * **Data is trouble** 1. Both groups spend lots of time dealing with this 2. Nowcasting 3. Low SNR 4. Very nonstationary * **Forecast evaluation is not settled** 1. WIS is equivalent to quantile loss, should use that. 2. Not optimized to predict turning points. 3. How do we create ensembles? ] .pull-right[ * **Hugely important to backtest properly** 1. Data is constantly revised 2. We see up to 10% "improvement" if we use finalized data 3. Bigger deal in the US * **Understanding the spatio-temporal dynamics is open** 1. How do "waves" propagate? 2. How important is mixing? 3. Effects of schooling? 4. Seasonality? * **True Counterfactual causal inference is open** * **Massive survey dataset** ] --- ## Delphi data  --- ## Many thanks * Jens, Dean, Dan, Sally and [BC COVID Modelling group](https://bccovid-19group.ca) * Jacob, Ryan, Rob, Larry, Valerie, Addison, Alden, Ken, Mike, Jed and the rest of [Delphi](https://delphi.cmu.edu/) * Shuyi, Wei (UBC Stat), Bryn (UBC Stat / BCCOVID) * Funding from NSERC, CANSSI * I get to benefit from the results of funding from Google, Facebook, Amazon, Change Healthcare, Quidel, SafeGraph, Qualtrics (you can too!) * Forecast evaluation from [Reich Lab](http://covid19forecasthub.org) and [Delphi](https://delphi.cmu.edu/forecast-eval/) <hr> -- .center[ **Questions** On these or other issues. ] --- class: middle, center, inverse # Extras --- <img src="index_files/figure-html/sim-sir-plot-1.svg" width="100%" />