Nonparametric estimation of time-varying reproduction numbers
Daniel J. McDonald

Mathematical modelling of disease / epidemics is very old
Daniel Bernoulli (1760) - studies inoculation against smallpox
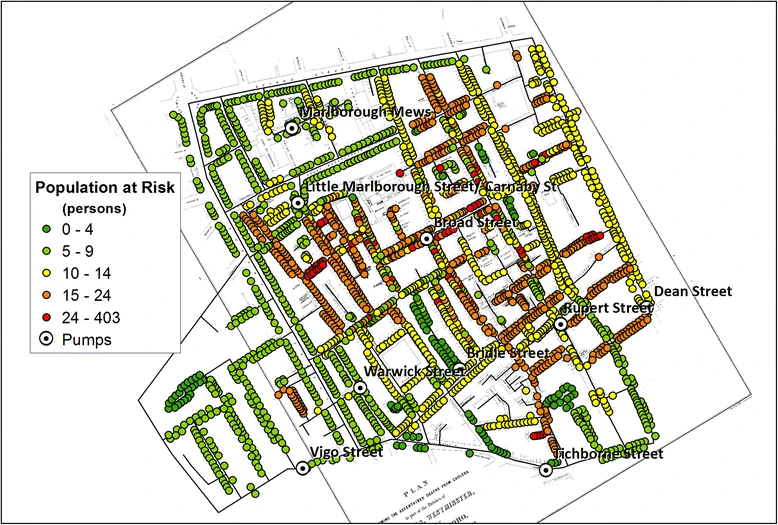
John Snow (1855) - cholera epidemic in London tied to a water pump
Ronald Ross (1902) - Nobel Prize in Medicine for work on malaria
Kermack and McKendrick (1927-1933) - basic epidemic (mathematical) model
\(R_0\) basic reproduction number
Dates at least to Alfred Lotka (1920s) and others (Feller, Blackwell, etc.)
The expected number of secondary infections due to a primary infection
- \(R_0 < 1\) ⟹ the epidemic will die out
- \(R_0 > 1\) ⟹ the epidemic will grow until everyone is infected

\(R_0\) is entirely retrospective
It’s a property of the pathogen in a fully susceptible (infinite) population
Each outbreak is like a new sample
To estimate something like this from data, the “bonehead” way is to
- Wait until the epidemic is over (no more infections circulating)
- Contact trace the primary infection responsible for each secondary infection
- Take a sample average of the number caused by each primary
- Possibly repeat over many outbreaks
30307-9/asset/daf522aa-ab16-4df8-ab69-212fb115a975/main.assets/gr4_lrg.jpg)
Of course no one actually does that
Lots of work on how to estimate \(R_0\)
Effective reproduction number
Suppose \(s\)% of the population is susceptible
Then, “the” effective reproduction number \(R=sR_0\)
Allows you to reason about things like
The level of vaccine coverage necessary to prevent an outbreak from growing uncontrollably.
So, for measles, if \(R_0\approx 15\), the disease will die out if immunity is
\[ sR_0 \leq 1 \Longrightarrow 1-s \leq 1-1/R_0 \approx 93\% \]
\(R(t)\) — instantaneous reproduction number
The effective reproduction number in the middle of an outbreak
Some of the population is immune, others are infected, others susceptible
The expected number of secondary infections at time \(t\) caused by an earlier primary infection
\(f(a) \geq 0,\ \forall a\) — the rate at which an infection of age \(a\) produces new infections
\[ \begin{aligned} R_0 &= \int_{0}^\infty f(a)\mathsf{d}a, \\ g(a) &= \frac{f(a)} {\int_{0}^\infty f(a)\mathsf{d}a} = f(a) / R_0. \end{aligned} \]
Can allow \(g(t, a)\), hold this fixed for now.
The generation interval distribution \(g(a)\)

Interlude on SIR models
Deterministic SIR
\[\begin{aligned} N &= S(0) + I(0) + R(0)\\ \frac{dS}{dt} & = -\frac{\beta}{N} S(t)I(t)\\ \frac{dI}{dt} & = \frac{\beta}{N} I(t)S(t) - \gamma I(t)\\ \frac{dR}{dt} & = \gamma I(t) \end{aligned}\]
\[R_0 = \beta/\gamma.\]
\[R(t) = \frac{\beta S(t)}{\gamma N}.\]
Stochastic SIR
\(R(t)\) and \(R_t\) — renewal equation
Alternatively: rather than working with a particular compartmental model
\(R(t)\) is defined implicitly through the renewal equation
\[ x(t) = R(t)\int_0^\infty x(t-a)g(a)\mathsf{d}a, \]
where \(x(t)\) are infections at time \(t\).
In discrete time,
\[ x_t = R_t\sum_{a=0}^\infty x_{t-a}\widetilde{g}_a = R_t (x * \widetilde{g}). \]
And stochasticly,
\[ \mathbb{E}\big[x_t\ |\ x_1,\ldots,x_{t-1}\big] = R_t\sum_{a=0}^\infty x_{t-a}\widetilde{g}_a = R_t (x * \widetilde{g}). \]
Most estimators start here: \[ \mathbb{E}\big[x_t\ |\ x_1,\ldots,x_{t-1}\big] = R_t\sum_{a=0}^\infty x_{t-a}\widetilde{g}_a. \]
- Assume \(\widetilde{g}\) is known
- Model \(x_t\ |\ x_1,\ldots,x_{t-1}\) as Poisson or Negative Binomial
- Turn some inferential crank
\(R_t\) for COVID-19 in the US
Source: US CDC Center for Forecasting Analytics
Estimating \(R_t\)
Data issues
\(x_t\) is Infections, but we don’t ever see those

- Replace infections with cases
- Replace generation interval with serial interval
- Assume we have the serial interval
Serial interval distribution
Standard model for \(R_t\)
\[ \begin{aligned} \eta_t &= \sum_{a=0}^\infty y_{t-a}p_a,\\ \\ y_t\ |\ y_1,\ldots,y_{t-1} &\sim \textrm{Poisson}(R_t\eta_t). \end{aligned} \]
- Using \(y\) instead of \(x\) to be cases or hospitalizations or deaths, incidence
- Using \(p\) for serial interval distribution (discretized)
- The MLE for \(R_t\) is just \(y_t / \eta_t\).
- This has really high variance, but unbiased.
- So everybody smooths it.
This isn’t going to give you “\(R_t\)”, because you didn’t use incident infections.
At least, the timing is wrong, because the observable happend after infection.
Deconvolve incidence first.
The state of the art (you saw some of these already)
{EpiEstim}(Cori, et al., 2013)
- Gamma prior on \(R_t\), but use a trailing window
- Super fast computationally
- Smoothness is ad hoc
{EpiFilter}(Parag, 2020)
- State space model
- One step smoothness: \(R_{s+1} \sim \textrm{Gaussian}(R_s,\ \alpha R_s)\)
- Uses a discretized particle filter-type algorithm
{EpiLPS}(Gressani, et al., 2022)
- Negative Binomial likelihood
- Smoothness via \(\log(R_t\eta_t) = \mathbf{B}_{t,:}\beta\)
- \(\mathbf{B}\) is cubic B-spline basis, weighted Ridge penalty on \(\beta\)
- More priors, use Metropolis Adjusted Langevin Algorithm
{EpiNow2}(CDC + CFA, Abbott, et al., 2023ish)
- Negative Binomial likelihood
- Smoothness via a GP prior
- Accommodates the sequence of delays from infection \(\longrightarrow\) ??
- Adjusts for real-time issues like partial reporting
- Big Bayesian MCMC in Stan. Very slow.
Our model
Let \(\theta_t := \log(R_t)\).
Use Poisson likelihood.
\[ \begin{aligned} \widehat{R} &= \exp(\widehat{\theta}) &\widehat{\theta} &= \mathop{\mathrm{argmin}}_\theta\; \eta^{\mathsf{T}}\exp(\theta) - \mathbf{y}^{\mathsf{T}}\theta + \lambda\Vert D^{(k+1)}\theta\Vert_1 \end{aligned} \]
- Convex, has a global optimum
- \(\lambda\) controls smoothness relative to data fidelity
- \(\ell_1\) penalty produces adaptive piecewise polynomials of order \(k+1\)
- Near minimax optimal for functions with bounded total variation
Local adaptivity — \(\ell_1\) vs. \(\ell_2\)
Polynomial order, \(k=0\)
\[
\begin{aligned}
D^{(1)} &= \begin{bmatrix}
1 & -1 & & & & \\
& 1 & -1 & & & \\
& & & \ddots && \\
& & & & 1 & -1
\end{bmatrix} \\ \\
&\in \mathbb{R}^{(n-1)\times n}
\end{aligned}
\]
Polynomial order, \(k=1\)
\[
\begin{aligned}
D^{(2)} &= \begin{bmatrix}
1 & -2 & 1 & & & \\
& 1 & -2 & 1 & & \\
& & & \ddots && \\
& & & 1 & -2 & 1
\end{bmatrix} \\ \\
&= D^{(1)}D^{(1)}\\ \\
&\in \mathbb{R}^{(n-k-1)\times n}
\end{aligned}
\]
Polynomial order, \(k=2\)
\[
\begin{aligned}
D^{(3)} &= \begin{bmatrix}
-1 & 3 & -3 & 1 & & \\
& -1 & 3 & -3 &1 & \\
& & & \ddots && \\
& & -1 & 3 & -3 & 1
\end{bmatrix} \\ \\
&= D^{(1)}D^{(2)}\\ \\
&\in \mathbb{R}^{(n-k-1)\times n}
\end{aligned}
\]
Estimation algorithm
\[ \mathop{\mathrm{minimize}}_\theta\; \eta^{\mathsf{T}}\exp(\theta) - \mathbf{y}^{\mathsf{T}}\theta + \lambda\Vert D^{(k+1)}\theta\Vert_1 \]
Estimation algorithm
\[ \mathop{\mathrm{minimize}}_{\theta,\ {\color{BurntOrange} \alpha}}\; \eta^{\mathsf{T}}\exp(\theta) - \mathbf{y}^{\mathsf{T}}\theta + \lambda\Vert D^{(1)}{\color{BurntOrange} \alpha}\Vert_1\quad {\color{BurntOrange} \textrm{subject to}\quad \alpha = D^{(k)}\theta} \]
Estimation algorithm
\[ \mathop{\mathrm{minimize}}_{\theta,\ \alpha}\; \eta^{\mathsf{T}}\exp(\theta) - \mathbf{y}^{\mathsf{T}}\theta + \lambda\Vert D^{(1)} \alpha\Vert_1\quad \textrm{subject to}\quad \alpha = D^{(k)}\theta \]
Alternating direction method of multipliers (ADMM)
\[ \begin{aligned} \theta &\longleftarrow \mathop{\mathrm{argmin}}_\theta\ \eta^{\mathsf{T}}\exp(\theta) - \mathbf{y}^{\mathsf{T}}\theta + \frac{\rho}{2}\Vert D^{(k)}\theta - \alpha + u \Vert_2^2 \\ \alpha &\longleftarrow \mathop{\mathrm{argmin}}_\alpha\ \lambda\Vert D^{(1)} \alpha \Vert_1 + \frac{\rho}{2}\Vert D^{(k)}\theta - \alpha + u \Vert_2^2 \\ u &\longleftarrow u + D^{(k)}\theta - \alpha \end{aligned} \]
Estimation algorithm
\[ \mathop{\mathrm{minimize}}_{\theta,\ \alpha}\; \eta^{\mathsf{T}}\exp(\theta) - \mathbf{y}^{\mathsf{T}}\theta + \lambda\Vert D^{(1)} \alpha\Vert_1\quad \textrm{subject to}\quad \alpha = D^{(k)}\theta \]
Alternating direction method of multipliers (ADMM)
\[ \begin{aligned} \theta &\longleftarrow {\color{Cerulean}\textrm{Proximal Newton / Fisher Scoring}} \\ \alpha &\longleftarrow {\color{BurntOrange}\textrm{Fused Lasso Signal Approximator}} \\ u &\longleftarrow u + D^{(k)}\theta - \alpha \end{aligned} \]
Solve sequentially for \(\Vert (D^{\dagger})^{\mathsf{T}}(\eta - y)\Vert_\infty = \lambda_1 > \cdots > \lambda_M=\epsilon \lambda_1\).
Results and features
Canadian Covid-19 cases
\(R_t\) for Canadian Covid-19 cases
Reconvolved Canadian Covid-19 cases
Example simulations for different methods
{rtestim} software

Guts are in C++ for speed
Lots of the usual S3 methods
Approximate “confidence” bands
\(\widehat{R}\) is a member of a function space
Arbitrary spacing of observations
Built-in cross validation
Time-varying delay distributions
{rtestim} software
Guts are in C++ for speed
Lots of the usual S3 methods
Approximate “confidence” bands
\(\widehat{R}\) is a member of a function space
Arbitrary spacing of observations
Built-in cross validation
Time-varying delay distributions
Approximation + Delta method gives
\[ \textrm{Var}(\widehat{R}) = \left(\textrm{diag}(\widehat{y}) + \lambda D^{\mathsf{T}}D\right)^{\dagger} \left(\frac{1}{\eta^2}\right) \]
{rtestim} software
Guts are in C++ for speed
Lots of the usual S3 methods
Approximate “confidence” bands
\(\widehat{R}\) is a member of a function space
Arbitrary spacing of observations
Built-in cross validation
Time-varying delay distributions
The solution is an element of the space of discrete splines of order \(k\) (Tibshirani, 2020)
- Lets us interpolate (and extrapolate) to off-observation points
- Lets us handle uneven spacing
{rtestim} software
Guts are in C++ for speed
Lots of the usual S3 methods
Approximate “confidence” bands
\(\widehat{R}\) is a member of a function space
Arbitrary spacing of observations
Built-in cross validation
Time-varying delay distributions
{rtestim} software
Summary and collaborators
Contributions
- Framework for \(R_t\) estimation
- Naturally imposes smoothness
- Fast, extensible, easy to use
- Has “confidence” bands
Future work
- Allow Negative Binomial likelihood
- Deconvolve incidence
- Forecast
- Mixed smoothness behaviours
- Figure out CV for convolution
- Better confidence bands
- Use relationship with growth rate
Collaborators and funding





{rtestim} — dajmcdon/rtestim-2025