flowchart LR
A("{epidatr}") --> C("{epiprocess}")
B("{epidatpy}") --> C
D("{covidcast}") --> C
C --> E("{epipredict}")
{epiprocess} and {epipredict}
R packages for signal processing and forecasting
Daniel J. McDonald, Ryan J. Tibshirani, and Logan C. Brooks
and CMU’s Delphi Group
CDC Flu Site Visit — 19 July 2023
Background
- Covid-19 Pandemic required quickly implementing forecasting systems.
- Basic processing—outlier detection, reporting issues, geographic granularity—implemented in parallel / error prone
- Data revisions complicate evaluation
- Simple models often outperformed complicated ones
- Custom software not easily adapted / improved by other groups
- Hard for public health actors to borrow / customize community techniques
Packages under development
{epiprocess}
Tools for signal processing
Basic processing operations and data structures
- General EDA for panel data
- Calculate rolling statistics
- Fill / impute gaps
- Examine correlations, growth rates
- Store revision history smartly
- Inspect revision patterns
- Find / correct outliers
epi_df: snapshot of a data set
- a tibble; requires columns
geo_valueandtime_value. - arbitrary additional columns containing measured values
- additional keys to index (
age_group,ethnicity, etc.)
epi_df
Represents a snapshot that contains the most up-to-date values of the signal variables, as of a given time.
epi_df: snapshot of a data set
Example data object documentation, license, attribution
case_death_rate_subset package:epipredict R Documentation
Subset of JHU daily state cases and deaths
Description:
This data source of confirmed COVID-19 cases and deaths is based
on reports made available by the Center for Systems Science and
Engineering at Johns Hopkins University. This example data ranges
from Dec 31, 2020 to Dec 31, 2021, and includes all states.
Usage:
case_death_rate_subset
Format:
A tibble with 20,496 rows and 4 variables:
geo_value the geographic value associated with each row of
measurements.
time_value the time value associated with each row of
measurements.
case_rate 7-day average signal of number of new confirmed COVID-19
cases per 100,000 population, daily
death_rate 7-day average signal of number of new confirmed deaths
due to COVID-19 per 100,000 population, daily
Source:
This object contains a modified part of the COVID-19 Data
Repository by the Center for Systems Science and Engineering
(CSSE) at Johns Hopkins University as republished in the COVIDcast
Epidata API. This data set is licensed under the terms of the
Creative Commons Attribution 4.0 International license by the
Johns Hopkins University on behalf of its Center for Systems
Science in Engineering. Copyright Johns Hopkins University 2020.
Modifications:
• From the COVIDcast Epidata API: These signals are taken
directly from the JHU CSSE COVID-19 GitHub repository without
changes. The 7-day average signals are computed by Delphi by
calculating moving averages of the preceding 7 days, so the
signal for June 7 is the average of the underlying data for
June 1 through 7, inclusive.
An `epi_df` object, 20,496 x 4 with metadata:
* geo_type = state
* time_type = day
* as_of = 2022-05-31 12:08:25.791826
# A tibble: 20,496 × 4
geo_value time_value case_rate death_rate
* <chr> <date> <dbl> <dbl>
1 ak 2020-12-31 35.9 0.158
2 al 2020-12-31 65.1 0.438
3 ar 2020-12-31 66.0 1.27
4 as 2020-12-31 0 0
5 az 2020-12-31 76.8 1.10
6 ca 2020-12-31 96.0 0.751
7 co 2020-12-31 35.8 0.649
8 ct 2020-12-31 52.1 0.819
9 dc 2020-12-31 31.0 0.601
10 de 2020-12-31 65.2 0.807
# ℹ 20,486 more rowsSliding examples on epi_df
Correlations at different lags
Outlier detection and correction
Sliding examples on epi_df
Growth rates
epi_archive: collection of epi_dfs
- full version history of a data set
- acts like a bunch of
epi_dfs — but stored compactly - allows similar funtionality as
epi_dfbut using only data that would have been available at the time
Revisions
Epidemiology data gets revised frequently. (Happens in Economics as well.)
- We may want to use the data as it looked in the past
- or we may want to examine the history of revisions.
Revision patterns
Example data object documentation, license, attribution
archive_cases_dv_subset package:epiprocess R Documentation
Subset of daily doctor visits and cases in archive format
Description:
This data source is based on information about outpatient visits,
provided to us by health system partners, and also contains
confirmed COVID-19 cases based on reports made available by the
Center for Systems Science and Engineering at Johns Hopkins
University. This example data ranges from June 1, 2020 to Dec 1,
2021, and is also limited to California, Florida, Texas, and New
York.
Usage:
archive_cases_dv_subset
Format:
An 'epi_archive' data format. The data table DT has 129,638 rows
and 5 columns:
geo_value the geographic value associated with each row of
measurements.
time_value the time value associated with each row of
measurements.
version the time value specifying the version for each row of
measurements.
percent_cli percentage of doctor’s visits with CLI (COVID-like
illness) computed from medical insurance claims
case_rate_7d_av 7-day average signal of number of new confirmed
deaths due to COVID-19 per 100,000 population, daily
Source:
This object contains a modified part of the COVID-19 Data
Repository by the Center for Systems Science and Engineering
(CSSE) at Johns Hopkins University as republished in the COVIDcast
Epidata API. This data set is licensed under the terms of the
Creative Commons Attribution 4.0 International license by Johns
Hopkins University on behalf of its Center for Systems Science in
Engineering. Copyright Johns Hopkins University 2020.
Modifications:
• From the COVIDcast Doctor Visits API: The signal
'percent_cli' is taken directly from the API without changes.
• From the COVIDcast Epidata API: 'case_rate_7d_av' signal was
computed by Delphi from the original JHU-CSSE data by
calculating moving averages of the preceding 7 days, so the
signal for June 7 is the average of the underlying data for
June 1 through 7, inclusive.
• Furthermore, the data is a subset of the full dataset, the
signal names slightly altered, and formatted into a tibble.
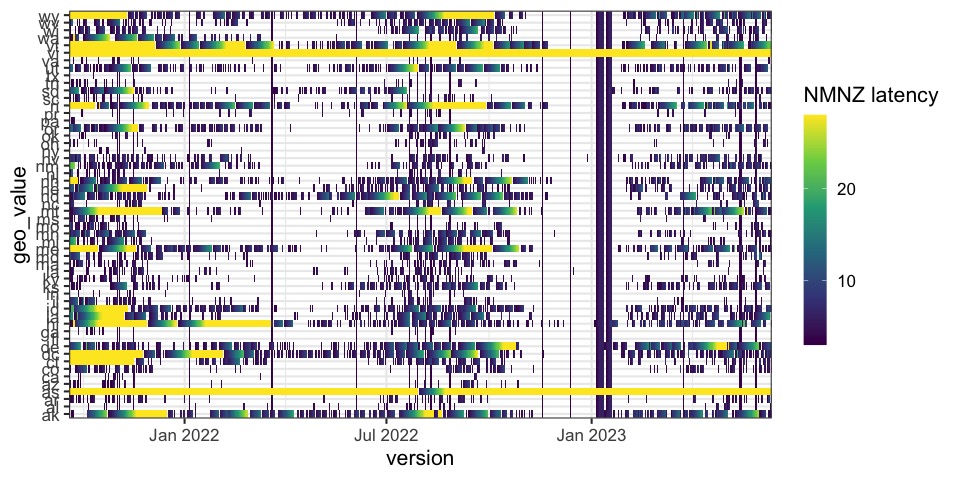
Latency investigation
How stale is my data over space and time?
There are (lots) of possible definitions.
- Nominal latency
-
the difference between the max
time_valueavailable for some location, as of some version - Nonmissing latency
-
nominal + recorded
NAs and skippedtime_values are latent - Nonmissing nonzero latency
- nonmissing + zeros are latent
- Nonmissing nonduplicate latency
-
nonmissing + duplications of the most recent non-
NAvalues are latent
Latency investigation
pos_first_true <- function(x) match(TRUE, rev(x), length(x) + 1) - 1
na0_latency <- function(dat, group_key, ref_time_value) {
group_by(dat, geo_value) %>%
complete(time_value = full_seq(time_value, period = 1L)) %>%
mutate(not_na0 = !(is.na(admissions) | admissions == 0L)) %>%
summarize(
nominal_latency = as.integer(ref_time_value - max(time_value)),
na0_latency = pos_first_true(not_na0) + nominal_latency
)
}
hhs_flu_hosp_archive %>%
epix_slide(na0_latency, before = 27, names_sep = NULL) %>%
rename(version = time_value)Latency investigation, > 2 days
{epipredict}
A forecasting framework
Philosophy of forecasting
We should build up modular components
Be able to add layers sequentially
- Preprocessor: do things to the data before model training
- Trainer: train a model on data, resulting in an object
- Predictor: make predictions, using a fitted model object
- Postprocessor: do things to the predictions before returning
A very specialized plug-in to {tidymodels}
{epipredict}
- Canned forecasters that work out-of-the-box
- Backtest using the versioned data
- Easily create features
- Modify / transform / calibrate forecasts
- Quickly pivot to new tasks
- Highly customizable for advanced users
Canned forecasters that work out of the box.
But, you can adjust a lot of options
We currently provide:
- Baseline flat-line forecaster
- Autoregressive-type forecaster
- Autoregressive-type classifier
- “Smooth” autoregressive-type forecaster
Canned forecasters that work out of the box.
But, you can adjust a lot of options
Example forecasting task
On day \(t\)
We want to predict
- new hospitalizations \(y\),
- \(h\) days ahead,
- at many locations \(j\).
We’re going to make a new forecast each week.
Canned forecasters that work out of the box.
But, you can adjust a lot of options
Flatline forecaster
For each location, predict \[\hat{y}_{j,\ t+h} = y_{j,\ t}\]
Canned forecasters that work out of the box.
But, you can adjust a lot of options
AR forecaster
Use an AR model with an extra feature, e.g.: \[\hat{y}_{j,\ t+h} = \mu + a_0 y_{j,\ t} + a_7 y_{j,\ t-7} + b_0 x_{j,\ t} + b_7 x_{j,\ t-7}\]
Basic autoregressive forecaster
- Predict
death_rate, 1 week ahead, with0,7,14day lags ofcasesanddeaths. - Use
lmfor estimation. Also create intervals.
The output is a model object that could be reused in the future, along with the predictions for 7 days from now.
Adjust lots of built-in options
rf <- arx_forecaster(
epi_data = edf,
outcome = "death_rate",
predictors = c("case_rate", "death_rate", "fb-survey"),
trainer = parsnip::rand_forest(mode = "regression"), # use {ranger}
args_list = arx_args_list(
ahead = 14, # 2-week horizon
lags = list(
case_rate = c(0:4, 7, 14),
death_rate = c(0, 7, 14),
`fb-survey` = c(0:7, 14)
),
levels = c(0.01, 0.025, 1:19/20, 0.975, 0.99), # 23 ForecastHub quantiles
quantile_by_key = "geo_value" # vary noise model by location
)
)Do (almost) anything manually
# A preprocessing "recipe" that turns raw data into features / response
r <- epi_recipe(edf) %>%
step_epi_lag(case_rate, lag = c(0, 1, 2, 3, 7, 14)) %>%
step_epi_lag(death_rate, lag = c(0, 7, 14)) %>%
step_epi_ahead(death_rate, ahead = 14) %>%
step_epi_naomit()
# A postprocessing routine describing what to do to the predictions
f <- frosting() %>%
layer_predict() %>%
layer_threshold(.pred, lower = 0) %>% # predictions / intervals should be non-negative
layer_add_target_date(target_date = max(edf$time_value) + 14) %>%
layer_add_forecast_date(forecast_date = max(edf$time_value))
# Bundle up the preprocessor, training engine, and postprocessor
# We use quantile regression
ewf <- epi_workflow(r, quantile_reg(tau = c(.1, .5, .9)), f)
# Fit it to data (we could fit this to ANY data that has the same format)
trained_ewf <- ewf %>% fit(edf)
# examines the recipe to determine what we need to make the prediction
latest <- get_test_data(r, edf)
# we could make predictions using the same model on ANY test data
preds <- trained_ewf %>% predict(new_data = latest)Visualize a result for 1 forecast date, 1 location
Code
fd <- as.Date("2021-11-30")
geos <- c("ut", "ca")
tedf <- edf %>% filter(time_value >= fd)
# use most recent 3 months for training
edf <- edf %>% filter(time_value < fd, time_value >= fd - 90L)
rec <- epi_recipe(edf) %>%
step_epi_lag(case_rate, lag = c(0, 7, 14, 21)) %>%
step_epi_lag(death_rate, lag = c(0, 7, 14)) %>%
step_epi_ahead(death_rate, ahead = 1:28)
f <- frosting() %>%
layer_predict() %>%
layer_unnest(.pred) %>%
layer_naomit(distn) %>%
layer_add_forecast_date() %>%
layer_threshold(distn)
ee <- smooth_quantile_reg(
tau = c(.1, .25, .5, .75, .9),
outcome_locations = 1:28,
degree = 3L
)
ewf <- epi_workflow(rec, ee, f)
the_fit <- ewf %>% fit(edf)
latest <- get_test_data(rec, edf, fill_locf = TRUE)
preds <- predict(the_fit, new_data = latest) %>%
mutate(forecast_date = fd, target_date = fd + ahead) %>%
select(geo_value, target_date, distn) %>%
pivot_quantiles(distn) %>%
filter(geo_value %in% geos)
ggplot(preds) +
geom_ribbon(aes(target_date, ymin = `0.1`, ymax = `0.9`),
fill = "cornflowerblue", alpha = .8) +
geom_ribbon(aes(target_date, ymin = `0.25`, ymax = `0.75`),
fill = "#00488E", alpha = .8) +
geom_line(data = bind_rows(tedf, edf) %>% filter(geo_value %in% geos),
aes(time_value, death_rate), size = 1.5) +
geom_line(aes(target_date, `0.5`), color = "orange", size = 1.5) +
geom_vline(xintercept = fd) +
facet_wrap(~geo_value) +
theme_bw(16) +
scale_x_date(name = "", date_labels = "%b %Y") +
ylab("Deaths per 100K inhabitants")Questions?

Packages for signal processing and forecasting